
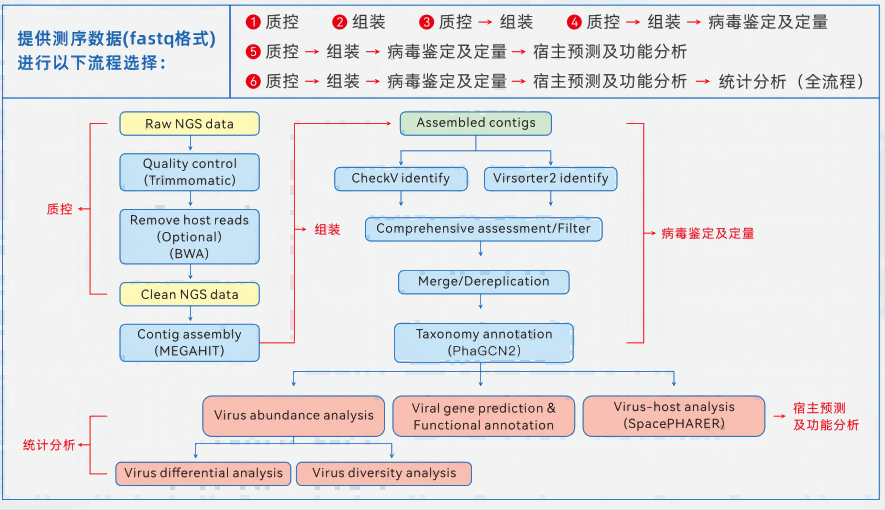
The macrovirome directly takes the genetic material of all viruses in the sample as the research object, and analyzes the virus community in it, involving human or animal blood, tissue, feces, plant tissues, as well as various complex environmental samples such as water and soil. Due to the small size of viral genomes, the relatively low content of viral nucleic acids in samples, the serious interference of host genome sequences, and the limited number and genomic information of known viruses, it is difficult to study metavirome from sample preparation to data analysis.
Nowadays, the development of large cohort samples and multi-omics produces massive data, and the pressure of data analysis and the overall project cycle are becoming more and more difficult to control. Cloud computing can effectively compress the analysis cycle through hundreds of sub-task parallel operations, combined with task scheduling and resource optimization and integration technologies, and greatly improve work efficiency and reliability.

Product Advantages:
l Step elastic self-selection, extreme cycle;
l Computing resources: a total of > 2,000,000 cores, which can be called at any time;
l Process quality: The process and database are updated every six months, and the scientific research results are reflected in Nature/Science/Cell/NC/PNAS;
l Service quality: omics and cloud computing expert team, point-to-point communication services.


Metagenomic sequencing is based on the whole microbial community in a specific environment as the research object, which gets rid of the technical limitations of microorganisms that are difficult to isolate and culture, and only needs to extract the total DNA of all microorganisms from the sample for sequencing.
Nowadays, the development of large cohort samples and multi-omics has led to a large amount of data output, so that the overall analysis cycle of the project is becoming more and more difficult to grasp.

Product Advantages:
l Step elastic self-selection, extreme cycle;
l Massive computing resources: a total of > 2,000,000 cores, which can be called at any time;
l Process quality: The process and database are updated every six months, and the scientific research results are reflected in Nature/Science/Cell/NC/PNAS;
l Service quality: omics and cloud computing expert team, point-to-point communication services.
